Everything to Know about the New WIPO Sequence Listing Standard – ST.26
A sequence listing is essential in all patent applications specifying nucleotide or amino acid sequences. The World Intellectual Property Organization (WIPO) has established international standards for preparing sequence listings. Thus, all relevant patent applications must comply with these guidelines or risk rejection of their patent application. WIPO recently rolled out its latest sequence listing standard – ST.26 – to improve data searching and transfer for patent offices, inventors, and applicants.
This article discusses ST.26, what is new in the ST.26 sequence listing standard, its benefits, the shortcomings of ST.25 standard, the difference between ST.25 and ST.26, and much more. But first, let us understand sequence listings and the role they play in patent applications.
Table of Contents
Introducing Sequence Listings
Inventions in the biotechnology field can include gene mutation, sequencing, regulation, expression, or silencing. As a result, a thorough explanation of the sequences is required in patent specifications to help examiners fully comprehend the innovation and compare it to prior art. As per the guidelines established by patent and trademark offices (PTOs), all DNA and protein sequences must be incorporated into the patent specifications. These sequences must also have a defined structure to ease comprehension. Sequence listing is the process of taking the raw sequences and inserting them into the necessary format in accordance with the rules of the patent office.
A gene is made up of a chain of polynucleotides composed of the amino acid codes, such as adenine (A), thymine (T)/uracil (U), cytosine (C), and guanine (G). The 20 naturally occurring amino acids are the building blocks of proteins. Each of these amino acids is represented by a unique one-letter code. Using “SEQ ID NO.” (for example, SEQ ID NO: 1), a sequence listing identifies the gene or protein and provides the whole sequence.
In case a patent application discloses nucleotide and/or amino acid sequences (in the description, claims, and/or drawings), the applicant must submit a sequence listing to the concerned intellectual property office (IPO). In general, the sequence listing must adhere to WIPO’s criteria concerning nucleotide and/or amino acid sequences disclosed in the patent application.
A sequence listing’s main objective is to make the sequence data searchable by intellectual property offices and the public. For instance, a number of patent offices, including the EPO (European Patent Office), KIPO (Korean Intellectual Property Office), and USPTO (United States Patent and Trademark Office), provide sequence data through publicly available databases, including EMBL (European Molecular Biology Laboratory), and NCBI (National Center for Biotechnology Information). A common international standard can help ensure hassle-free transfers and searching of data between and on such databases.
Sequence listings solve a critical problem that inventors and patent offices often face. The next section of this article delves into it.
Need for Sequence Listing
A sequence listing offers a systematic way to compile all biological sequence information provided in a patent application into one document. It facilitates the recording and transmission of biological sequence data from patent applications to searchable databases utilized by public and national patent offices. Therefore, it is crucial to include a sequence listing that satisfies the standard requirements outlined by international organizations, regardless of whether they were created synthetically, organically, or artificially.
Biological sequences serve as references and prior art for future studies and inventions. Sharing this data in a standardized format makes it easier to compile and incorporate biological sequence data in searchable databases. A sequence listing offers a standardized way to present all the biological sequence information disclosed in a patent application in one document. Most particularly, it provides a list of the nucleotide (RNA or DNA) and/or amino acid protein sequences mentioned in a patent application by enumerating their residues that satisfy sequence length thresholds.
Till recently, all sequence listings in patent applications had to comply with WIPO’s ST.25 sequence listing standard. Patent applicants used software, such as PatentIn and BiSSAP, for preparing sequence listings that are ST.25-compliant, and delivered the same to the IPO on paper or in .txt format. The new ST.26 sequence listing standard is now in place, and all sequence listings must adhere to the new guidelines established. Moreover, the sequence listing must also comply with other WIPO guidelines to further accelerate hassle-free comprehension and data transfer. Failure to meet the sequence listing standards can lead to the rejection of a patent application.
The purpose of the new sequence listing standard is to harmonize the way nucleotide and amino acid sequence listings are submitted in patent applications around the world. In accordance with the requirements, the applicant can create a single sequence listing application that will be accepted by all receiving offices, such as international searching and preliminary examination authorities for the international phase and all designated and elected offices for the national phase.
It strives to make sequence listings easier to present and publicize for the benefit of applicants, the general public, and examiners. An international standard also facilitates sequence searching and eases the exchange of sequence data in electronic form. It also aims to improve the accuracy and benchmark of nucleotide and amino acid sequences provided in international applications.
Before getting familiar with the new standard, it is important to understand the previous sequence listing norms. The next section delves into the same.
ST.25 Sequence Listing Standard
The term “ST.25” refers to the international standard that was adopted in 2009. This standard specified how nucleotide and amino acid sequences should be presented in a sequence listing. The WIPO ST.25 standard served as the basis for the US sequence regulations (37 CFR 1.821–1.825). Sequence listings in ST.25 format are required for any applications needing it and having a filing date or international filing date prior to July 1, 2022.
The fields in an ST.25 sequence listing marked from 110 through 170 are commonly referred to as the “header” fields. These header fields, which apply to all the sequences in the sequence listing, are only included once in the beginning. As sequence listings are frequently filed as separate documents from the specification of a patent application, the header fields are used to associate sequence data in the sequence listing with the relevant patent application.
A numbered sequence identifier is given to each sequence in an ST.25 sequence listing. The sequence identifiers start with “1” and increase sequentially by integers. Each sequence’s genetic identification can be found in the <210> field and the first line of the <400> field. Sequences are identified in a patent application description, claims, or drawings by their respective sequence identifiers, which are denoted by the prefix “SEQ ID NO:” from the sequence listing. The WIPO ST.25 standard was followed until recently in sequence listing filings.
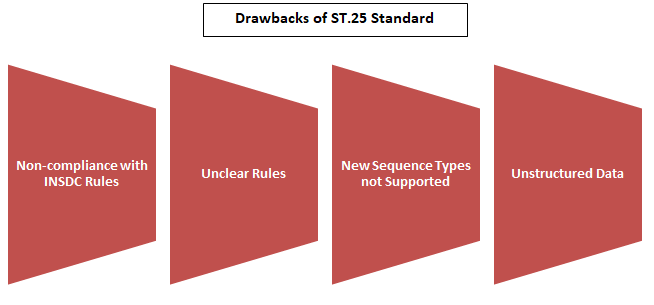
Drawbacks of ST.25 Sequence Listing Standard
Following are some of the major issues with the ST.25 standard that necessitated the introduction of ST.26:
- The format prescribed by ST.25 did not meet the INSDC (International Nucleotide Sequence Database Collaboration) specifications, and therefore, resulted in the loss of data when imported into public databases.
- Additionally, several ST.25 regulations needed more clarity, causing IPOs worldwide to apply and interpret them differently.
- Data was unstructured, and the ST.25 format was challenging to employ for automated validation and data exchange.
- Another issue with the ST.25 standard was that it did not cover the types of sequences that are popular and widely used these days, such as nucleotide analogs, D-amino acids, and branched sequences. This made it impossible to find these sequences in searchable databases. In addition, the information could not be verified as it was unstructured.
These shortcomings in the ST.25 prompted the development of the ST.26 sequence listing standard. The primary objective of the new guidelines is to standardize and enhance the filing requirements for sequence listings and make it easier to search for nucleotide and amino acid sequences associated with a patent application. The next section discusses the ST.26 sequence listing standard in great detail.
ST.26 – The New Sequence Listing Standard
“ST.26” refers to an international standard that outlines how nucleotide and amino acid sequences are to be displayed in a sequence listing using an XML format. It was adopted in October 2021 and came into effect on July 1, 2022. The WIPO ST.26 sequence listing standard serves as the basis for the US sequence regulations (37 CFR 1.831–1.835). The standard requires sequence listings in the XML format for all applications that have a filing date or international filing date on or after July 1, 2022.
A single file in XML 1.0 format, encoded with Unicode UTF-8, must be used to present an ST.26 sequence listing. Additionally, it must adhere to the WIPO Standard ST.26 Document Type Definition (DTD).
An ST.26 sequence listing in XML format consists of two basic sections:
- A general information section that contains the application’s bibliographic information, including the applicant’s name, the inventor’s name, the application number, the date of application filing, the invention title, and the earliest priority application.
- A sequence data part that contains one or more sequences as well as every feature identifying those sequences.
The MPEP (Manual of Patent Examining Procedure) updates to address WIPO Standard ST.26 are forthcoming.
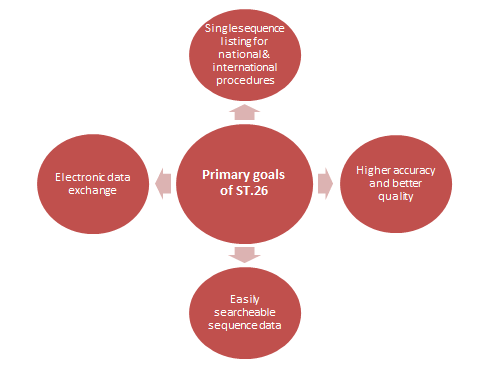
The purpose of ST.26 is to standardize sequence listing procedures across all national patent offices, taking into account improvements in biotechnology, and adhering to international regulations for sequence databases. The INSDC specification aims to create a single sequence listing format that is accepted globally and compatible with other international databases.
In contrast to the “plain text” TXT format prescribed by ST.25, sequence listings must be presented in a single file using XML format. The adoption of sequence data in XML format is meant to make it easier to search for data, especially in public databases. The main features of ST.26 are as follows:
- Applicants can file a single sequence listing for patent applications that can be used for both international and national or regional procedures.
- The public, examiners, and applicants will all benefit if sequences are presented more accurately and in higher quality.
- It will make it easier to search listings in sequence data.
- The new standard will facilitate electronic data sharing and database entry for sequence information.
Launch of ST.26 Sequence Listing Standard
The new WIPO ST.26 standard came into effect on July 1, 2022. Therefore, all patent applications disclosing nucleotide and/or amino acid sequences must now include a sequence listing that is in line with this new standard. The INSDC databases, including GenBank and EMBL-EBI, can now accept more sequence data in electronic form due to WIPO’s new ST.26 standard for sequence listings. Thus, it will be simpler to create and submit a sequence listing under the revised rules, which can help applicants save time and money. These rules will also make accessing and comparing sequence listings between patent and scientific publications simpler, thus enhancing the quality of search results.
The next section sheds light on all the changes made and new rules introduced by the ST.26 standard.
What’s New in ST.26 Sequence Listing Standard?
The ST.26 standard has introduced various new features to sequence listings. Below are some of the most significant ones:
- Applicants must now share sequence listings in XML format in contrast to TXT or PDF under ST.25.
- It improves the depiction of sequences (due to the structure of the sequence listing in XML format), supports automation of data verification, and streamlines IPO procedures.
- Another significant change is that now a single applicant name and inventor name can be submitted for the general information contained in the sequence listing. In any event, this involves a formal modification rather than a substantive one.
- When it comes to priority information, only the oldest priority is necessary. Furthermore, multiple titles of the invention may also be submitted, each in a different language.
- It requires the sequence to be uniquely designated as DNA, RNA, or AA, coupled with a mandatory qualification to identify the molecule more accurately.
- Additionally, unlike the ST.25 sequence listing rules, which only required L-amino acids to be included, the new guidelines now mandate the incorporation of D-amino acids, nucleotides, branched sequences, as well as analogs.
- Furthermore, it permits the adoption of a single sequence listing across the globe, barring any necessary language translations.
- It also specifies which sequence disclosures must be included in a sequence listing, which ones are allowed, and how these sequences must be represented.
- Small sequences are also prohibited under ST.26 standard.
Difference Between ST.25 and ST.26 Sequence Listing Standards
The major differences between ST.25 and ST.26 are listed in the table below. It also emphasizes how ST.26 standard has simplified the patent application.
| ST.25 Standard | ST.26 Standard |
| ASCII text in numerical identifier format. | UTF-8 (Unicode)-encoded XML format with components and attributes. |
| Not necessary to mention: D-amino acids Linear sections of branched sequences Nucleotide analogs | Must have: D-amino acids Linear sections of branched sequences Nucleotide analogs |
| Sequence annotation: Only feature keys | Sequence annotation: Key features as well as qualifiers |
| Sequences are allowed to include: < 10 specifically defined nucleotides < 4 specifically defined amino acids | Unacceptable sequences: < 10 specifically defined nucleotides< 4 specifically defined amino acids Note: Any nucleotide and amino acid other than “n” and “X” is regarded as being “specifically specified.” |
| Possible to incorporate all priority applications. | Only the most important application can be submitted. |
| The names of all applicants and inventors may be listed. | There can only be one applicant name and potentially one inventor name. This should be the first/primary applicant or innovator. |
| Only one invention title can be mentioned. | It is possible to include several invention titles in different languages. |
| Normal Latin characters must be used to write the inventors’ names and the names of their inventions. | Invention titles can contain any valid Unicode character, and inventor names can contain any valid Unicode character combined with a basic Latin translation. |
| Sequences are only categorized as DNA, RNA, or PRT. | Sequences are classified as DNA, RNA, or AA and as the mandatory qualifier “mol type” to further specify the type of molecule. |
| Organism names: Latin genus/speciesVirus name“Artificial sequence” “Unknown” | Organism names: Latin genus/speciesVirus name“Synthetic construct”“Unidentified” |
| “u” stands for uracil in nucleotide sequences. | The symbol “u” is not a recognized nucleotide symbol. In DNA and RNA sequences, the letter “t” stands for thymine and uracil, respectively. Modified nucleotide bases uracil in DNA and thymine in RNA must be represented by the symbol “t” in ST.26, and described in a feature table using the feature key “modified base.” |
| Three-letter acronyms are used to represent amino acid sequences. | One-letter acronym is used to represent an amino acid sequence. |
| The definition of “n” and “Xaa” variable residues must be provided in a feature. | A default value is assumed for “n” and “X” variable residues with no definitions. A definition is essential when “n” or “X” represent residues other than the default value. |
| The format for feature locations is not clearly defined. | Strictly defined formats for feature location; allows usage of “<” and “>” in all sequence types as well as “^,” “join,” “order,” and “complement” in nucleotide sequences. |
| Nucleotide sequence and amino acid translation, known as “mixed mode” sequence, is acceptable. | “Mixed mode” sequences are not permitted, but a “translation” qualifier could include a translation of a nucleotide. |
Major Features of ST.26 Sequence Listing Standard
The major features of ST.26 that apply to all sequences, including nucleotide and amino acid sequences, are listed below:
- Each sequence ought to have its own unique sequence identification number. Without a series, the identification number must only start at one and rise in integers. For a sequence identification number, a triple zero representing a deliberately skipped sequence must be used in its place.
- The two sections of a sequence listing are the general information section and the sequence data section. Additionally, a document-type declaration must be used to provide the sequence listing as a single XML file. The general information section includes bibliographic data that is only used to associate the sequence listing with the patent application for which it is submitted.
- There are one or more sequence data elements in the sequence data section, each containing details about a single sequence.
- Additionally, the INSDC and UniProt specifications must be followed when creating the sequence data elements.
There are tools that can help patent applicants prepare ST.26-compliant sequence listing, and among the most popular of these is WIPO Sequence Suite. Let us learn more about this software.
Putting ST.26 Standard into Practice with WIPO Sequence Listing Software
Any XML file editing tool that complies with WIPO ST.26 standard may be used to construct a sequence listing as per the new rules. However, due to ST.26 standard’s complexity, experts strongly advise utilizing software specifically created for the purpose, such as the WIPO Sequence Suite. Member states asked WIPO to create this common tool for all offices and applicants alike. As a result, WIPO Sequence Suite was built in partnership with patent offices worldwide to make it easier to create, verify, and author ST.26-compliant sequence listings.
There are two options in the WIPO sequence listing software—import project and import sequence listings—that allow you to upload additional files and continue working on them. The sequence listing verification feature helps users evaluate their sequence listings. The project name of the application files can be found under the project header. Here, all previous projects will be displayed, and one can easily continue working on any of these.
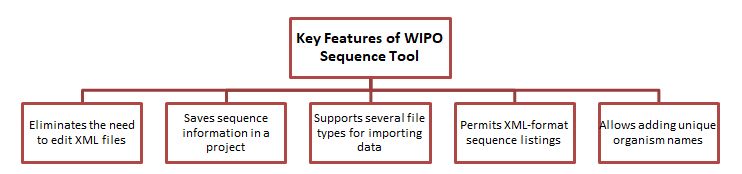
WIPO Sequence Suite also boasts a user-friendly interface which facilitates creating ST.26-compliant XML format sequence listings. Furthermore, it allows users to input data from ST.25 sequence listings, ST.26 sequence listings projects, multi-sequence format files, and other types of files. One may also prepare a sequence listing simply by entering a name and adding the sequences. The software can also validate sequence listings in XML format. Thus, using the WIPO sequence tool, an applicant can:
- Recognize and save a project-specific application and sequence data.
- Add feature keys and qualifiers to sequences by choosing from simple drop-down choices.
- Verify project data and produce an XML sequence listing that is compliant with the latest guidelines.
- Check the accuracy of an existing XML sequence listing.
- Create “human-readable” project data for simple review.
- Save unique applicant and inventor details.
- Record unique organism names.
- Import data from several file types, including ST.25 sequence listings, ST.26 sequence listings, ST.26 projects, raw files, multi-sequence format, and FASTA files.
- Facilitate the translation of free text qualifier values.
The WIPO Sequence Validator is an online service that helps IPOs confirm submitted listings’ adherence to the WIPO ST.26 standard. The application’s main objective is to give intellectual property offices a web service to validate XML files in WIPO ST.26 format and make sure they are compliant with WIPO ST.26 sequence listing standard. The Committee on WIPO Standards oversaw the development of these tools in collaboration with patent offices all over the world.
The tool consumes files from a local file system and generates a verification report with validation results and, optionally, returns the results of the validation process, i.e., verification report, by calling an endpoint.
When converting ST.25 sequence listing to ST.26 project, it is essential to keep the following in mind:
- The imported ST.25 sequence listing should be legitimate and adhere to all the ST.25 requirements. Data loss and other unanticipated complications may occur from importing an erroneous ST.25 sequence listing into the WIPO sequence tool.
- When an ST.25 sequence listing is imported, an ST.26 project, and not an ST.26 XML format sequence listing, is created. An ST.25 sequence listing does not contain certain mandatory elements, such as the “mol type” qualifier, which is present in an ST.26 sequence listing. A genuine ST.26 XML sequence listing cannot be produced until users add more data to the resulting ST.26 project.
- It is crucial that users carefully analyze each note in the detailed “Import Report” generated by the WIPO sequence tool upon importing ST.25 text files to comprehend what data modifications have taken place and take any necessary precautions to prevent data loss.
New Sequence Listing Inclusion Criteria in ST.26
There are several inclusion requirements for sequence listings in ST.26 that were not included in ST.25 standard. For instance, unconventional sequence types, such as D-amino acids, nucleotide analogs, and branched sequences, are specifically required in the ST.26 standard.
Furthermore, ST.26 has modified the sequence listings’ minimum length requirement. Also, sequence listings can only include unbranched sequences and linear sections of branched sequences that contain:
- 10 or more precisely defined nucleotides, or
- 4 or more precisely defined amino acids.
Sequences that are too short need to be stated elsewhere in the patent specification, not in a sequence listing. One significant difference between ST.25 and ST.26 is that the latter clearly restricts the inclusion of sequences that are shorter than the minimum requirement.
Moreover, only “specifically defined” residues count towards the length as per the ST.26 standard. Here, “specifically defined” means any nucleotide other than those denoted by the symbol “n,” and any amino acid other than those denoted by the symbol “X” – listed in Annex I. To put it another way, the sequence length does not include nucleotides denoted by “n” and amino acids denoted by “X.” For instance, a nucleotide sequence represented by 5’-acgtnnacgt-3’ is not allowed to be listed in a sequence since it only contains eight clearly defined nucleotides.
Annex I also provides different symbols for other combinations of alternative nucleotides. For instance, “v” is the symbol for “a or c or g; not t/u”, and “b” is the symbol for “c or g or t/u; not a.” Thus, the term “specifically defined,” in the context of a nucleotide sequence, excludes the symbol “n,” and includes different symbols defining other combinations of alternative nucleotides.
We can see how using the new ST.26 sequence listing standard can be extremely beneficial for various parties involved. In the following section, we will go over its benefits in great detail.
Advantage of ST.26 Sequence Listing Standard
With the new sequence listing standard ST.26, data verification automation has improved, IPO procedures have been made simpler, and the sequences are presented more clearly (due to the XML formatted sequence listing structure). There are numerous other advantages, some of which are listed below:
- According to the new standard, applicants may provide a single sequence listing in a patent application for regional, national, or international purposes.
- ST.26 will improve the precision and standard of sequence presentations.
- It will make sequence data searching much easier than before.
- The new sequence listing standard will enable the electronic sharing of sequence data and its incorporation into digital databases.
- ST.26 will ensure that IP offices are in agreement.
- It will speed up IP offices’ processing and increase the automation of data validation.
- The XML sequence listings’ format will lead to an improvement in submission quality.
When to Create the Sequence Listing Document?
The timeline for creating a sequence listing document is a crucial factor for applicants. In addition to the paper copy required in the office, a computer-readable copy of the sequence listing must also be submitted with the patent application. The data shared in this machine-readable format must be identical to the printed sequence listing.
The sequence listing document should ideally be created concurrently with the patent application, allowing room for modifications at a later stage. The SEQ ID NOs must remain the same in the sequence listing document and the draft of the patent application to facilitate necessary modifications in both formats. Nevertheless, it is advisable to produce the two documents simultaneously.
Best Practices for Smooth Transition into ST.26 Standard
The following is a list of some of the recommended practices for a smooth transition into ST.26 sequence listings:
- The ST.25 standard will still be in effect for a PCT application submitted before July 1, 2022, even if it enters the EP or GB phase later. However, all new applications, including new applications claiming priority from applications submitted previously and new European divisional applications filed after July 1, 2022, must follow the ST.26 standard.
- An ST.25 sequence listing may need conversion into the ST.26 format for a new application. For instance, the EPO has confirmed that the conversion of the ST.25 sequence listing into an ST.26 sequence listing, and a statement that the new sequence listing does not add subject matter beyond the parent application, which is descended from a patent application submitted before July 1, 2022, are required for divisional applications submitted on or after July 1, 2022. On the other hand, the UKIPO mandates that the sequence listings that go with new GB divisional applications be provided in the same manner as the parent application.
- When creating new ST.26-compliant listings or converting an existing ST.25 listing to the ST.26 format, applicants should be aware that more time may be needed, especially for listings that contain sequences affected by revisions, such as:
- Sequences with D-amino acids or nucleotide analogs
- RNA or DNA containing the nucleotide thymine
- Branched sequences
- Sequences with additional feature annotations
- Furthermore, when converting ST.25 sequence listings to ST.26-compliant listings for applications claiming an earlier priority date or European divisional applications, extreme caution should be exercised to avoid adding or removing subject matter. Given the EPO’s strong stance on the new subject matter and priority entitlement, this could be a significant problem if the converted ST.26 sequence listing is filed here.
Why Choose Sagacious IP for Sequence Listing?
Sagacious IP is one of the most well-known and largest intellectual property service providers globally. Our patent experts can create sequence listings that comply with the most recent guidelines of national and international IP offices. Apart from that, Sagacious IP is the ideal sequence listing partner for a variety of reasons, including:
1. Experts with experience in Creating Compliant Sequence Listing
- We have a team of biotechnology and biological sciences experts who are conversant with WIPO, USA, and other jurisdictional norms.
- All our experts receive extensive training in creating listings that adhere to the latest standards.
- The team consults with the client and provides any additional information needed for preparing sequence listing as required by the patent and trademark office (PTO).
- We constantly integrate in-house algorithms with tools for preparing sequence listing, such as WIPO Sequence Suite, PatentIn 3.5, and BiSSAP software, to minimize efforts and generate results that are specific to the client.
2. Preparing Error-free PTO-Compliant Sequence Listings
In the past year, Sagacious IP has created more than 300+ error-free sequence listings in PTO-acceptable format.
3. Low Cost and Quick Turnaround Time
We provide quick turnaround time and cost-effective sequence listing services.
Our Success Stories
Sagacious IP has a team of experts who can create flawless sequence listings for your patent applications. In the past few years, we have helped numerous clients prepare flawless sequence listings and obtain IP protection. Here are some such case studies.
Case Study 1:
One of our clients wanted us to prepare a sequence listing of approx. 1400 sequences in ST.26 format for filing an international application. The client needed the sequence listing in 6-8 business days. Hence, to meet WIPO Standard ST.26 sequence listing requirements, Sagacious IP assigned multiple resources and prepared files in XML and TXT formats using the WIPO Sequence 2.2.0 tool. The modified sequences majorly included 2-prime O methylation of nucleotides and 2-prime-fluoro modifications of nucleotides. Accordingly, we were able to prepare and deliver the sequence listing timely.
Case Study 2:
One of our regular clients for sequence listing wanted us to convert ST.25 to ST.26 format. However, the ST.25 listings required other modifications as per the ST.26 sequence listing rules. The client intended to file a European divisional application and wanted to be sure that the new listing met the appropriate ST.26 requirements.
Sagacious IP understood the requirement well and converted the ST.25 sequence listing into ST.26 format. Later the modifications were described as per ST.26 sequence listing rules, where phosphorothioate bonds using x^y locators were defined along with 2-prime O methylation of nucleotides and 2-prime-fluoro modifications of nucleotides. Hence, Sagacious IP prepared, generated and validated sequence listing containing modified sequences as per ST.26 rules and delivered the listing timely.
Case Study 3:
In another case, a client approached Sagacious IP to correct a defective sequence listing. The listing did not comply with 37 CFR 1.831(b), which states that every relevant patent application must contain a “Sequence Listing XML.” After understanding the client’s requirements, Sagacious IP prepared the sequence listing in XML format, in line with WIPO ST.26 sequence listing standard requirements, and was timely delivered within one to two business days.
Case Study 4:
In another case, a client approached Sagacious IP to convert ST.25 listing containing 13000+ sequences into ST.26 format. Sagacious IP imported the ST.25 listing into the WIPO tool to prepare XML files in ST.26 format and validated the same.
Conclusion
Any new application that includes sequence listing must now adhere to WIPO’s ST.26 sequence listing standard. ST.26 is a significant improvement on ST.25, and applicants must make sure that their applications satisfy all the additional guidelines established through this new standard. Even though XML and TXT documents have significantly different appearances, the new WIPO sequence program enables applicants to create ST.26-compliant sequence listings and examine a more understandable form of the sequence listing. WIPO Sequence Suite automatically satisfies many of the new ST.26 standards; thus, applicants or agents who intend to file applications containing sequence listings are encouraged to acquire and become familiar with them.
Sagacious IP’s experts can help you create error-free sequence listings for your national and international patent applications. Our team has several years of experience in the field and can swiftly create ST.26-compliant sequence listings at an affordable price. We have delivered more than 25000 projects for over 5000 clients in 100+ countries and 16+ languages. You can read more about your expertise and experience on the sequence listing service page.
– Pooja Chhikara (Life Sciences & Chemistry) and the Editorial team
Having Queries? Contact Us Now!
"*" indicates required fields




