ST26 Sequence Listing Standard: Easing the Access to Sequence Data
All patent applications disclosing nucleotide and/or amino acid sequences must include sequence listings as per international filing standards. A new sequence listing standard – ST26 – is coming into force from July 1, 2022, that is expected to improve the data structure for automatic data exchange and verification between patent offices and biological sequence search databases. Although ST26 is a WIPO (World Intellectual Property Organization) initiative, the decision on how accessible the new sequence listings on public portals should be is in the hands of individual patent offices. The need for a new listing standard arose as the current standard, ST25, did not support automated validation and data exchange.
This article will talk about the difference between ST25 and ST26, the shortcomings of ST25, the changes brought in by WIPO in the new standard, its benefits, and much more. But first, let us have a clear understanding of what exactly sequence listing is.
Table of Contents
What is Sequence Listing?
The sequence listing is a part of the description of patent applications which includes nucleotide and/or amino acid sequence(s). It is a prerequisite for patents in the biotech space, such as inventions related to biomarkers, antibodies, oligonucleotides, etc. The sequence listings provided in patent applications must comply with the prescribed filing regulations to avoid any hindrance in the patent approval process. The sequence listing standard currently in practice is ST25.
Now that we have understood what sequence listing is, let us get to know the sequence listing-related rules that one must follow while submitting a patent application in the biotech domain.
Patent Rules regarding Sequence Listings
Here are some critical rules pertaining to sequence listings in patent applications:
- Inventors must file sequence listing in computer-readable text format alongside every patent application, disclosing a sequence of nucleotides or amino acids.
- If the international application discloses one or more nucleotide or amino acid sequences that are not furnished in computer-readable text format, then the Searching Authority would send a notice to the applicant to submit the same. The applicant also has to pay the late furnishing fee as specified in the Fifth Schedule within one month. In case the applicant fails to comply with the notice, the Searching Authority will proceed and search the international application without the sequence listing to a reasonable extent.
As mentioned earlier, a new sequence listing standard is coming into force from July 1, 2022. So, next, we will focus on the need for ST26.
Need for a New Sequence Listing Standard
The need for a new sequence listing standard stems from the shortcomings of the standard in current practice. These are mentioned below.
- ST25 format did not comply with INSDC (International Nucleotide Sequence Database Collaboration) requirements.
- The rules were unclear, and IP offices worldwide interpreted and enforced the laws differently.
- ST25 rules did not cover sequence types that are common today (e.g., nucleotide analogs, D-amino acids, and branched sequences).
- The data was unstructured.
- ST25 format was challenging to use for automated validation and data exchange.
Now, let us understand the changes introduced through the new standard.
Sequence Listing Changes Introduced through ST26
Key changes brought by WIPO through ST26 are mentioned in figure 1 below:
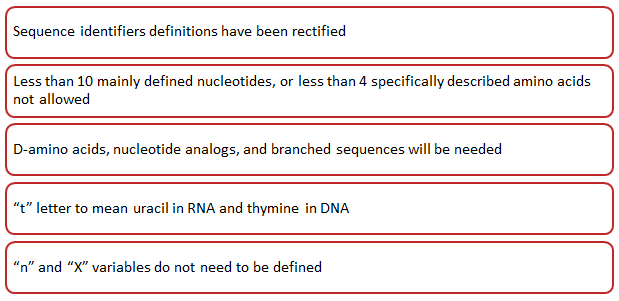
Given below are some more changes made by WIPO through ST26.
- .xml file types will be required to tackle data loss, and support exchange of data readable by humans as well as machines.
- Further sequence annotation options will be available.
- The names and options of the organisms will be updated.
- Instead of single-letter codes, three-letter codes will be preferred.
- The format of the feature location has been modified.
- Mixed-mode sequences will not be permitted anymore.
- The latest priority information (instead of all) can be incorporated.
- Some more straightforward alterations regarding the language requisitions have also been made.
To enhance the understanding of the old and new sequence listing standards, let us highlight the key difference between the two.
How does ST26 differ from ST25?
Here’s how ST26 has made the patent application simpler.
| Contents | ST25 Standard | ST26 Standard |
| File Format | ASCII.txt with numeric identifiers | XML with elements and attributes |
| Permitted Sequence | Permitted to include sequences: < 10 specifically defined nucleotides < 4 specifically defined amino acids | Prohibited sequences: < 10 specifically defined nucleotides< 4 specifically defined amino acids |
| Applicant/Inventor | All applicant and inventor names can be inculcated | Only one applicant and optionally one innovator can be incorporated |
| Annotation | Annotation of sequences: Feature keys only | Annotation of sequences: Feature keys and qualifiers |
| D-amino acid/branched sequences/analogs | Not required to include: D-amino acidsLinear portions of branched sequencesNucleotide analogs | Must include: D-amino acidsLinear portions of branched sequencesNucleotide analogs |
| Priority application | All priority application info may be included | Only the earliest priority application info can be included |
| Title | One invention title permitted | Multiple invention titles permitted, each one in a different language |
| Applicant/inventor names | The name and invention titles of the applicant/inventor must be in basic Latin characters | The name of the applicant/inventor may be included using any valid Unicode character along with a basic Latin translation or transliteration |
| Type of Sequence | Sequences identified as RNA, DNA, or PRT only | Sequences identified as RNA, DNA, or AA along with a mandatory molecule type qualifier to further describe the molecule For RNA: Genomic RNA, mRNA, RNA, rRNA, transcribed RNA For DNA: Genomic DNA, unassigned DNA For AA: Protein |
| Uracil Symbol | ”u” represents uracil in nucleotide sequences | “t” represents thymine in DNA sequences and uracil in RNA sequences |
| Organism Name | Latin genus/species Virus name “Artificial sequence” “unknown” | Latin genus/species Virus name “Synthetic construct” “unidentified” |
| Variables | “n” and “Xaa” variables must have a definition provided in a feature | Default value assumed for “n” and “X” variables with no definition |
| AA Seq. Presentation | Amino acids sequences represented by three letter abbreviations | Amino acids sequences represented by one letter abbreviation |
| Mixed Mode Sequence | “Mixed mode” sequences permitted – nucleotide sequence with amino acid translation shown below | No “Mixed mode”; nucleotide translations are included in “translation” qualifiers only |
| Feature Location | Feature location format not clearly defined | Clearly defined feature location formats; permits use of “” in all sequence types, and “^”, “join”, “order”, and “complement” in nucleotide sequences |
Benefits of ST26 Sequence Listing Standard
The many benefits of the ST26 standard are mentioned below.
- The new standard will permit applicants to create a single sequence listing in a patent application for international or national purposes, or regional procedures.
- ST26 will enhance the accuracy and quality of the presentation of sequences.
- It will facilitate the searching of sequence data.
- It will allow sequence data to be exchanged electronically and introduced into computerized databases.
- ST26 will ensure agreement among IP offices.
- It will boost the automation of data validation and streamline processing by IP offices.
- Submission quality will improve due to the structure of XML sequence listings.
Preparing ST26-Compliant Sequence Listings
One can use the WIPO Sequence tool to convert old ST25 TXT sequence listings into the new ST26 XML format. This responsibility of sharing sequence listings as per the new standard rests with the applicant, as the Patent Office does not perform the conversion. One can also use WIPO Sequence to check for errors in the ST26 sequence and view the new standard’s XML file in a human-readable format in the browser.
About WIPO Sequence
WIPO Sequence is a global software tool that allows patent applicants to prepare amino acid and nucleotide sequence listings compliant with the WIPO ST26 standard as a part of a national or international patent application. Here are some key features of WIPO Sequence:
- The tool developed by WIPO supports authoring, validation, and generation of ST26 compliant sequence listings.
- It eliminates the need to edit XML files.
- One can save sequence information in a project, validate it, and generate a sequence listing in the ST26-compliant format.
- It supports importing data from ST26 sequence listings, ST26 projects, ST25 sequence listings, multi-sequence format files, raw format files, and FASTA format files.
- The tool allows validation of sequence listings in XML format.
- It supports the export and import of XLIFF files used by translators.
Conclusion
Patent applications in the biotech domain require sequence listing in a standard format as prescribed by WIPO. Previously, the applicants used to follow the ST25 standard, whereas from July 1, 2022, a new standard, ST26, will come into force. The purpose of shifting from ST25 to ST26 is to enhance presentations accuracy and quality, facilitate sequence data searching, and allow the sequence data to be exchanged electronically.
Failing to comply with WIPO’s sequence listing standards can result in the rejection of a patent application. Sagacious IP’s team of patent experts can help you prepare sequence listing compliant with the latest guidelines. Visit our sequence listing service page to know more.
– Pooja Chhikara (Life Sciences) and the Editorial Team
Having Queries? Contact Us Now!
"*" indicates required fields




