Learn About Sequence Listing & Best Practices to Prepare Error Free Sequence Listings
Table of Contents
Key-points covered in the webinar (Learn About Sequence Listing & Best Practices to Prepare Error Free Sequence Listings)
- What is sequence listing and why is it needed?
- Introduction to patent office standards for sequence listing.
- How to prepare error-free sequence listings.
- Introduction to the tool for preparing sequence listing.
- Case studies on how Sagacious IP helped attorneys in preparing error-free sequence listing.
Key-note Speakers
Pooja Chhikara, Assistant Manager LSC, Sagacious IP
Manisha Singh, Project Manager LSC, Sagacious IP
Submit Your Information to watch the Webinar Video:
"*" indicates required fields
Webinar Transcript:
Pooja Chhikara – Learn about Sequence Listing & Best Practices to Prepare Error-Free Sequence Listings
Before I go on to introduce this topic, I am delighted to welcome all the participants from different regions including the United States, Europe, India, China, and other Asian countries. Your participation is a wonderful encouragement to the efforts and attempt that we are making to raise awareness and spread knowledge that has been honed by Sagacious IP over several years of working with Inventors, R&D Organizations, IP Departments, and IP Law Practices.
Along with me, I have Manisha Rani, Project Manager in the Life Sciences team. She holds more than four years of experience in the patent domain, skilled in the conducting landscape, FTO, Invalidity and Prior-Art Searches, and proficient in assessing IP in various fields such as Biotechnology, Microbiology, Immunology, etc.
Welcome to the Webinar, Manisha.
Manisha Rani – Hey Pooja, thanks for having me on the webinar.
Pooja Chihikara: The other speaker of the session is me. I’m a qualified patent agent, having more than five years of experience in several projects in patent analysis, competency in identifying and assessing IP in biotechnology, microbiology, biochemistry, pharmaceutical, and immunology. It’s a pleasure to be on the webinar.
Sequence Listings
Before we start off with the presentation today, let me ask Manisha for her initial remarks on today’s webinar topic i.e., Learn about Sequence Listing & Best Practices to Prepare Error-Free Sequence Listings.
Manisha Rani – First of all, a very warm welcome to all the participants of the webinar!
First, we begin why the sequence listing is important as lots of patent filing is happening in the biotechnology domain and biological sequences are an important part of biotic invention. It brings unique challenges for their effective and equal use in practice, since the function can only be determined with computer aided technology, for example, Bioinformatics.
Hence, the forms in which sequences are disclosed matter greatly. Similarly, the scope of patent rights requires computer readable data and tools for comparison critically. The primary data provided at the time of filing must be comprehensive. It should be standardized timely and meaningfully. We can say Sequence listing plays an important role during filing of a patent.
Pooja Chhikara – Thanks, Manisha, for setting the context of this webinar. The participants of the webinar can drop us an email later at [email protected], and we will answer to that as well.
Let us now get started with the main part of the presentation and for that, let me invite Manisha to start with the webinar.
Over to you, Manisha!
What Is Sequence Listing?
Manisha Rani – Thank you once again, Pooja. Let’s begin with “What Is Sequence Listing?”
As biotechnology invention involves gene mutation, gene sequences and regulating or silencing of gene in the domain. Invention and advancements such as antibodies, genetically modified organism, and biomarkers involve sequencing of genes, amino acid, and nucleotide bases to transform the genetic make-up and to produce modified (enhanced) living organism.
As we talk about gene mutation and gene silencing, we should be aware of what gene is.
Basically, gene is a chain of polynucleotide which is composed of four bases named Adenine, which is represented by A, Thymine represented by T, Guanine by G, and Cytosine by C. They all code for amino acids.
During filing of such patent application that contain nucleotides and amino acid sequencing, they all need to be entered into the patent specification, and it’s necessary to have a particular format or you can say structure instead of the raw format. Basically all this process of entering these row sequences in the required format as per the guidelines of Patent Office is known as sequence listing.
Patent Office Requirements for Sequence Listing
Now, let’s discuss what are the Patent Office Requirements for Sequence Listing?
The number one is all the sequences provided in the patent application are to comply with the PTO filing regulation.
The second thing, all the filing should be done in a standard format that is, according to the US law, which is US CFR 1.821-1.825, and according to WIPO standard format, that is ST. 25, which is the standard for presentation of nucleotide and amino acid sequence listing in international Patent Application under the PCT.
All the DNA sequences having 10 or more nucleotide bases and all unbranched known amino acid sequence with four or more amino acid should be listed. Or we can say that there are at least 10 specifically defined nucleotides or four specifically defined amino acids.
The sequence listing rule is applicable to all sequence present in a patent application irrespective of that the sequence is present in claims or in description. So basically we can say that all sequences should be listed either they are presented in claims or in description, but the priority should be given to the claims sequences.
All the sequence listings recognize the gene or protein by “SEQ ID NO:” (for example SEQ ID NO: 1)
The sequence ID number is unique for each sequence.
Advantages of Sequence Listing
Now we move forward and let’s discuss about what the advantages of sequence listing are.
Within a patent document, genetic sequences are viewed both legally and practically. That is either chemical compounds or as information-encoding elements. The sequence structure creates importance within the context of patent eligibility or infringement issues.
So sequence listing allows the applicant to create a single listing that is acceptable to all filing offices and receiving offices. It also allows the simpler presentation of sequence, which is easy to understand. It also increases the accuracy of amino acids and/or of nucleotide sequence, which makes it easy to understand by the public.
Tools for Preparing Sequence Listing
Now let’s move forward and talk what tools are required for preparing sequence listings. Basically we have different tools for listing sequences.
PatentIn
The first is PatentIn. The version is 3.5.1 or earlier. We have two versions for this, but it is advisable to use a more advanced version. PatentIn is computer design software which helps in the preparation of a patent application containing nucleic acid and amino acid sequences.
It accepts data about the sequence, it validates the data, and then it creates a sequence listing file. This software has a worldwide applicability. This software includes different modules like a sequence editor. This is basically the primary tool within PatentIn. It enables us to enter and modify nucleic acids and protein sequences listing, as well as it imports patented generated ST.25 sequence listing file. We can say that sequence data files created by another editor or word processor. They are stored as ASCII i.e., American Standard Code for Information Interchange (text file). It also includes a sequence generator.
For example, we have entered all the data necessary for our patent application. It enables us to generate the application, and the application consists of a computer readable ST.25 compliant file containing all the sequence listing file.
BiSSAP
We have another tool for preparing sequence listing i.e., BiSSAP. We can use any version that is 1.3 or earlier one, but it is advisable to use the advanced version. This is also a computer program which is designed to facilitate the creation of sequence listing for patent applications containing biological sequences.
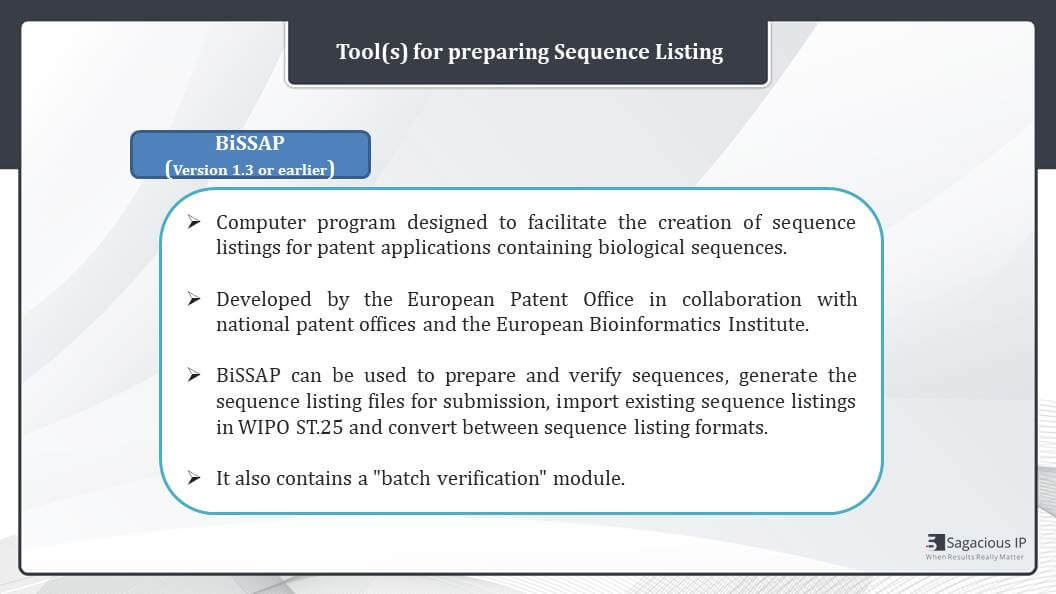
The software is developed by European Patent Office in collaboration with National Patent Office and European Bioinformatics Institute. It can be used to prepare and verify sequences. It generates a sequence listing file for submission, and you can import existing sequence listing in WIPO standard 25. And it also helps to convert between sequence listing formats that are WIPO ST.25 and XML proposal. It also contains a batch verification module.
Now, over to your Pooja, so that you can explain the exact module and methodology for the performing sequence listing.
Thank you.
Key Consideration During Filing/Drafting
Pooja Chhikara – Thank you, Manisha, for the elaborated introduction of the topic. So moving forward, let me start with the mentioning of important considerations while filing or drafting a patent application comprising biological sequences.
Features Applicable to All Sequences
The first is the claim sequences should be given priority and should start with sequence ID No: 1. Next, the sequence listings shall be presented as a separate part of the description, be placed at the end of the description, and the page should be entitled “Sequence Listing”. These listings should begin on a new page and have independent page numbering. Next, how the sequences should be presented – Each sequence shall be assigned to a separate sequence identifier that is a Sequence ID NO. The sequence identifiers shall begin with one and increase sequentially by integers.
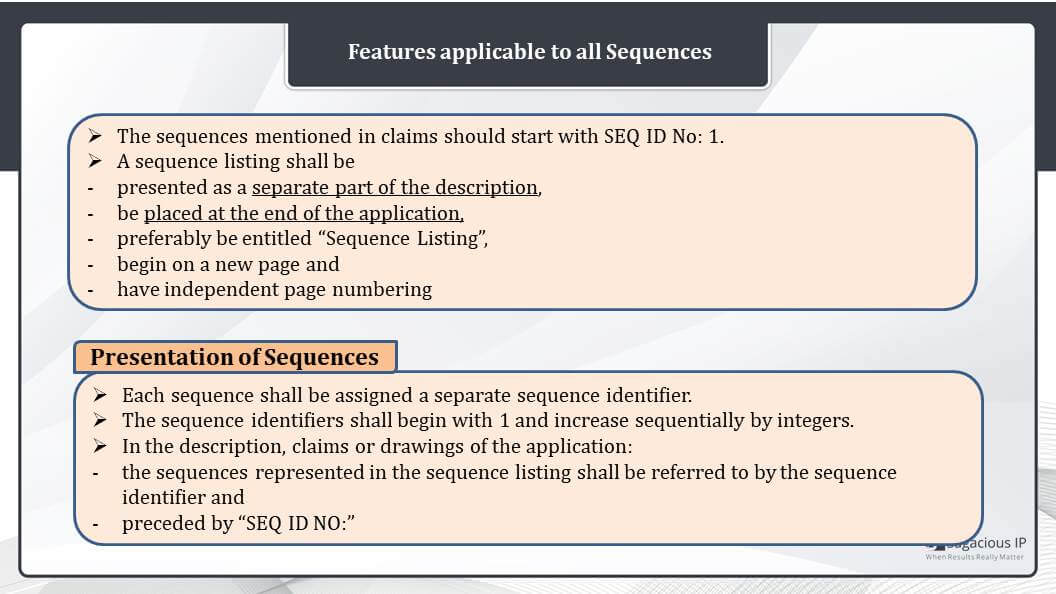
For example, SEQ ID NO: 1, then next should be SEQ ID NO: 2, and like that.
Sequences present anywhere in the description, claims or drawing of the patent application. They shall be referred to by the sequence identifier preceded by “SEQ ID NO:”
Nucleotide Sequences
How the nucleotide sequences are presented – any nucleotide sequence shall be presented only by a single strand in 5’-end (5 prime ends) to 3’-end (3 prime ends) direction from left to right. The strand from 3’-end to 5’-end shall not be represented in this sequence. We just have to provide one strand.
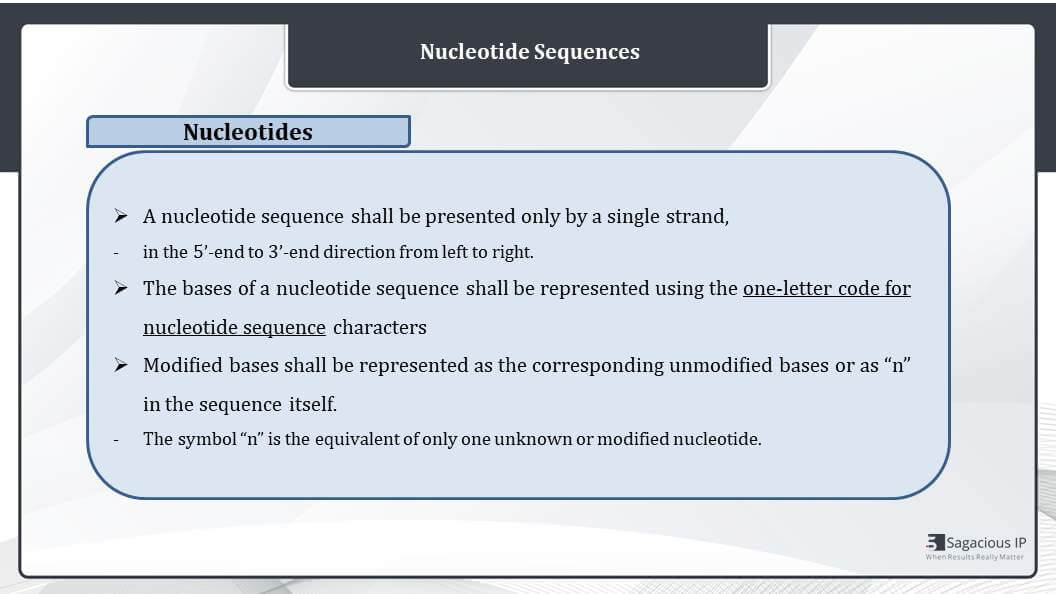
The nucleotide bases of a sequence shall be presented, using the one-letter code for nucleotide sequence characters. For example, A, T, G, and C are four nucleotides.
Next, all the modified bases shall be represented as the corresponding unmodified bases as defined by the ST.25 document, or it can be represented in the sequence itself. The symbol “n” is the equivalent of only one unknown or modified nucleotide, and the modifications shall be further described in the feature section of the sequence listing.
Format to Be Used
Talking about the format of nucleotide sequences, a nucleotide sequence shall be listed with a maximum of 60 bases per line with a space between each group of 10 bases. That means there would be a gap after every set of 10 nucleotide bases, and these sequences be listed in group of 10 bases except in the coding parts of the sequence.
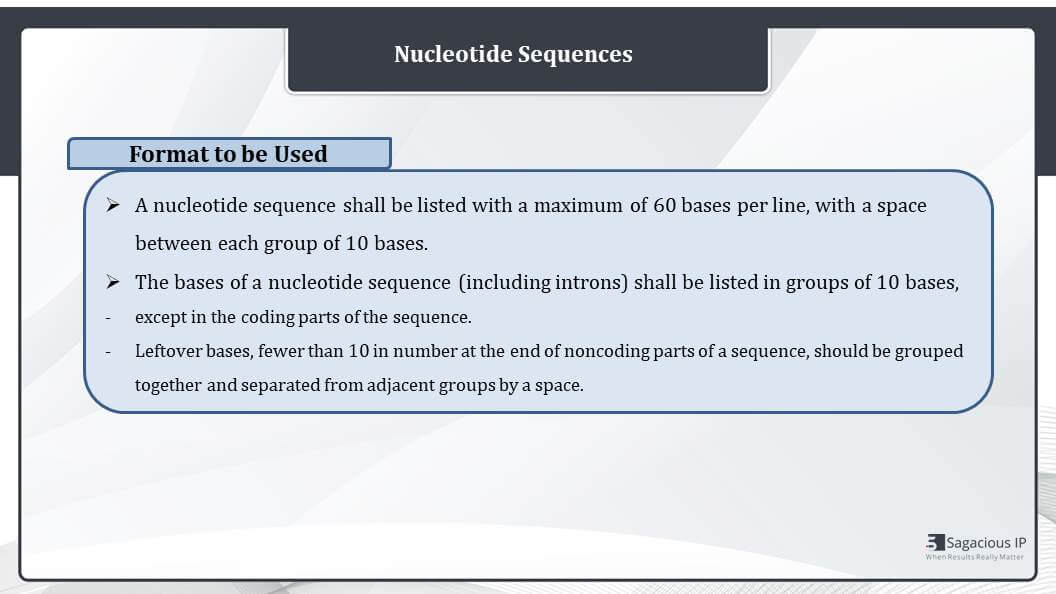
Further, the leftover bases that are fewer than 10 in number, at the end of the noncoding parts of the sequence, they should be grouped together and separated from adjacent groups by a space.
Protein Sequences
Amino Acids
Then, how are the amino acids sequences presented? The amino acids in a protein sequence, they shall be listed in the amino to carboxy direction from left to right, and here also we provide the one single strand. The amino and carboxy groups shall not be represented in the sequence.
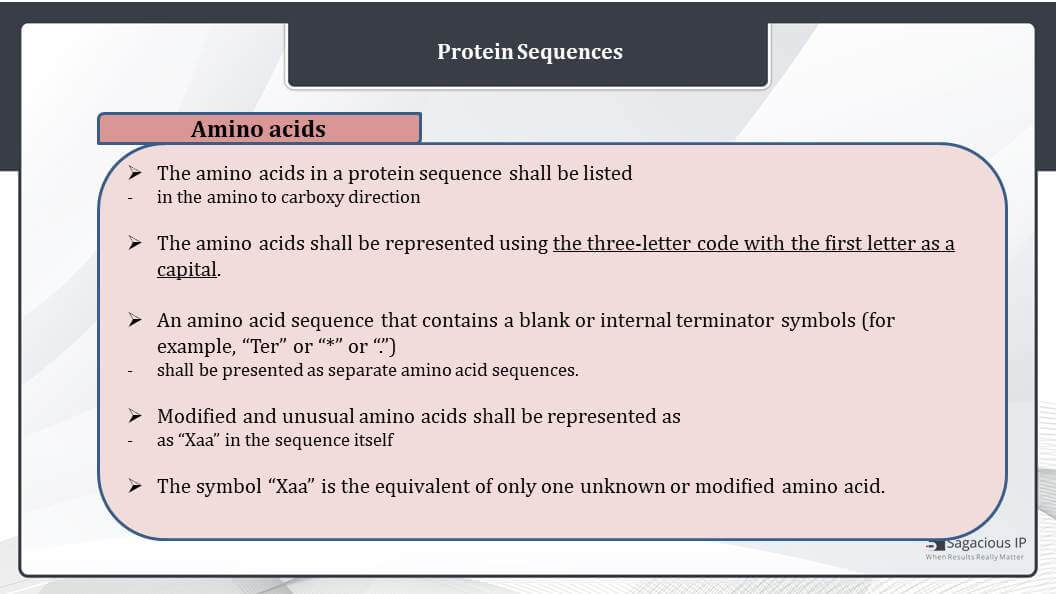
The amino acids shall be represented using the three-letter code with the first letter as a capital. We all are aware that the amino acids can be represented by a single-letter code and a three-letter code. In sequence listing, we only use the three-letter code to represent the amino acid sequences.
In amino acid sequence that contains a blank or internal terminator symbols, for example, “Ter” or a “*” or a “.”, or dot. They shall be represented as a separate amino sequence. It may not be represented as a single amino acid sequence as there is a break in this sequence. This should be presented as two different sequences.
The modified and usual amino acids shall be represented as the corresponding unmodified amino acids per ST.25 document or as “Xaa” in the sequence itself, and the modification shall be further described in the feature section of the sequence listing.
The symbol “Xaa” is the equivalent of only one unknown or modified amino acid, and it is similar to the one that we discussed for nucleotides. We used “N” for nucleotides and Xaa for amino acids to define unknown or modified bases.
Format to Be Used
Further a protein or peptide sequence shall be listed with a maximum of 16 amino acids per line with a space provided between each amino acid. That means there will be a gap after every three-letter code of amino acid.

Next, the amino acids corresponding to the codons in the coding part of a nucleotide sequence, they shall be placed immediately under the corresponding codons, where a codon is split by an intron. The amino acid symbol should be given below the portion of the codons containing two nucleotides.
When Should the Sequence Listing Document Be Prepared
Now, apart from the presentation of the sequence, another important consideration is the timeline for preparing a sequence listing document. A copy of the sequence listing must be submitted in computer readable form at the time of filing of the patent application. In addition to the paper copy that is required in the office.
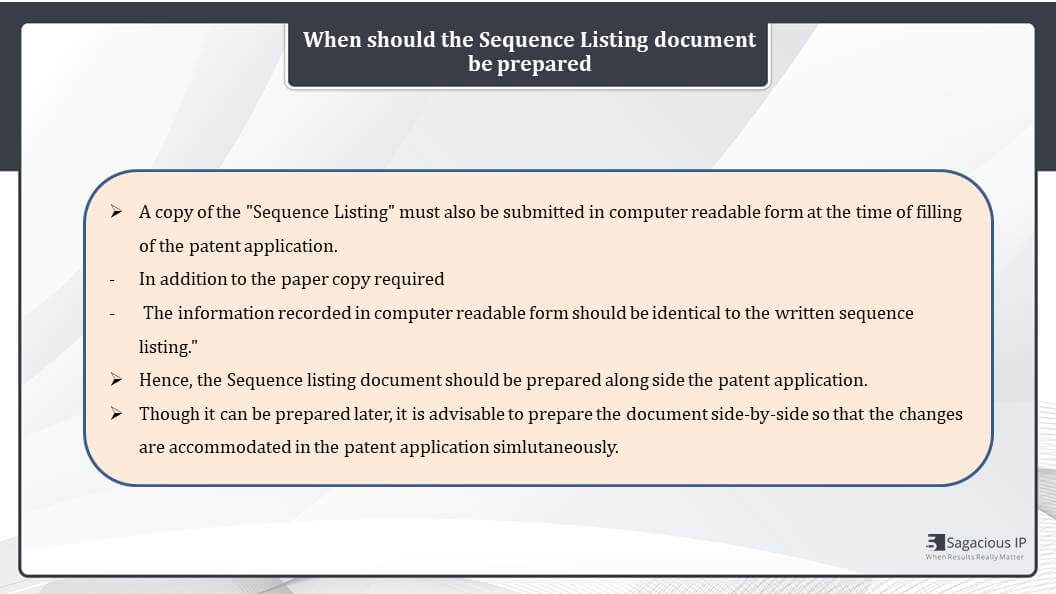
The information recorded in this computer readable form should be identical to the written sequence listing. Ideally the sequence listing document should be prepared alongside the patent application so we can prepare it later as well. But it is advisable that we prepare it side by side because we have to keep the SEQ ID NOs same in the sequence listing document and the draft of patent application, so we can make the changes accordingly in both the documents.
Methodology for Sequence Listing
Moving forward, let’s briefly understand how a sequence listing document is prepared using PatentIn.
PatentIn Interface
This is the PatentIn software interface. This is the home page of PatentIin. You can see various steps like Project, Application steps, and Organism – this organism name for the sequence. Then, there’s an “Add” for adding a sequence and “Import” for working or modifying a previous sequence file. We can import a previously made sequence listing file and then can modify it further.
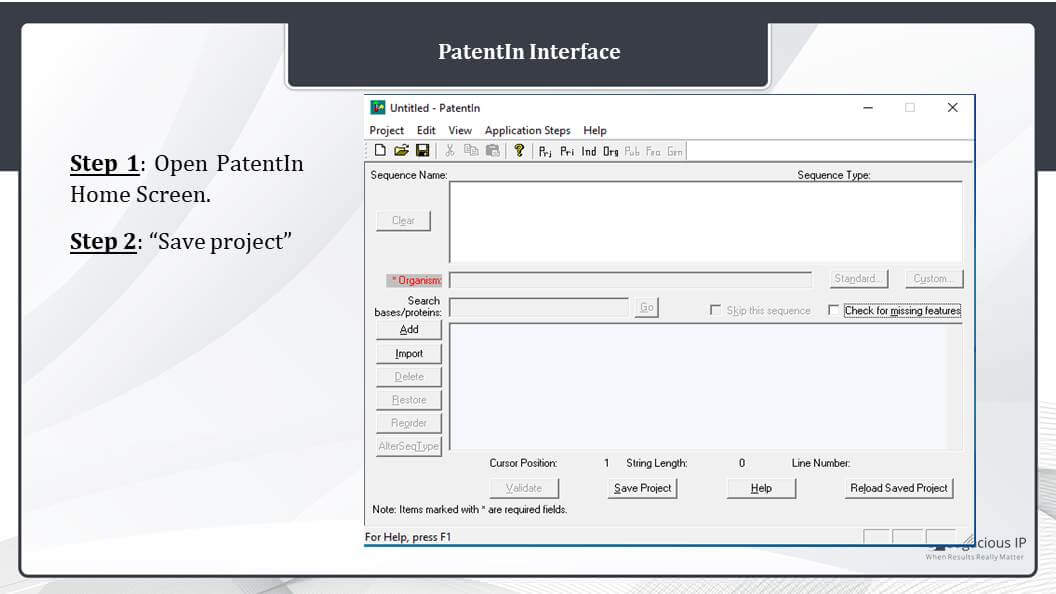
Talking about the methodology, the first obvious step for preparing a sequence of listing document is opening of PatentIn home screen. The second step is saving the project by clicking on the Save Project. We can name the file according to our requirement.
Application Data
The third step is entering all the application data in the tab “Application Step”. Here the first option is Project Data. Project Data, Title of Invention, and Application File Reference are the mandatory fields, and we have to enter these details. Then, we can press okay, after entering them.
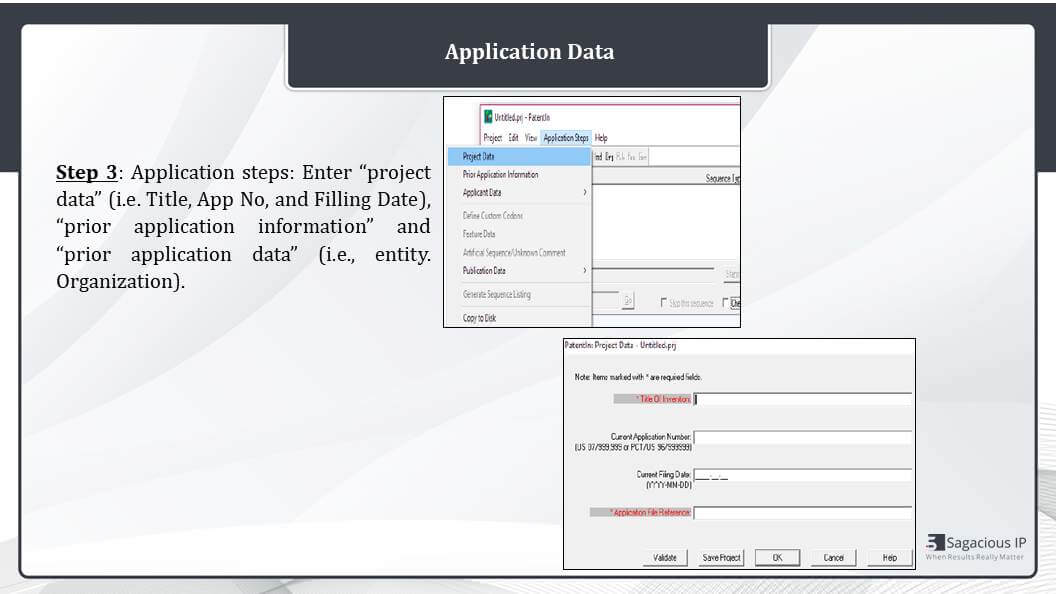
The next header is prior application information where the information regarding the priority application is provided. Then, in application data, we enter the inventor and assignee information, and then we can move ahead with entering the sequences.
Adding Sequence
The next step includes addition of sequence type for example, DNA, and RNA. First of all, we click on the add button, and then we can select the DNA, RNA or protein as required by clicking on the add tab. And here in the upper tab, we need to add Sequence Numbers like 1 or 2, or 1, or SEQ ID NO: 1 – like that.
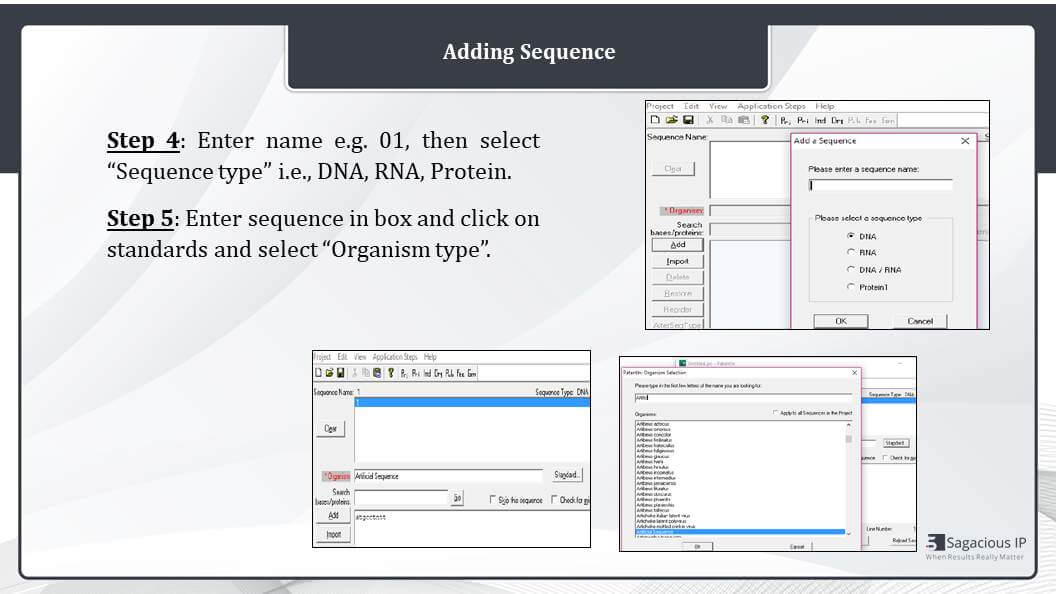
Once the sequence is selected and the number is provided, we need to enter the nucleotide or protein sequence that need to be entered here in this box, like we have entered our sample here ATGC. We have entered this. We should add the organism name which can be selected from the dropdown box. Now, once we click on it or drop down box would appear and we can select the organism.
Comment for Artificial Sequence
In case the organism is not known and it is an artificial sequence, then we must add a comment for the artificial sequence. So in case of the artificial sequence, entering your comment is must. The moment artificial sequence is selected; you will see a pop up here, which is where we need to add a comment. And this artificial comment can be later changed by clicking on the application steps and selecting the artificial sequence or unknown comment.
So for writing the comment, we use free text in the use of free text shall be limited to a few short terms indispensable for the understanding of the sequence, and it shall not exceed four lines with a maximum of 65 characters per line for each given data element when written in English. And any further information shall be included in the main part of the description of application in the patent application.
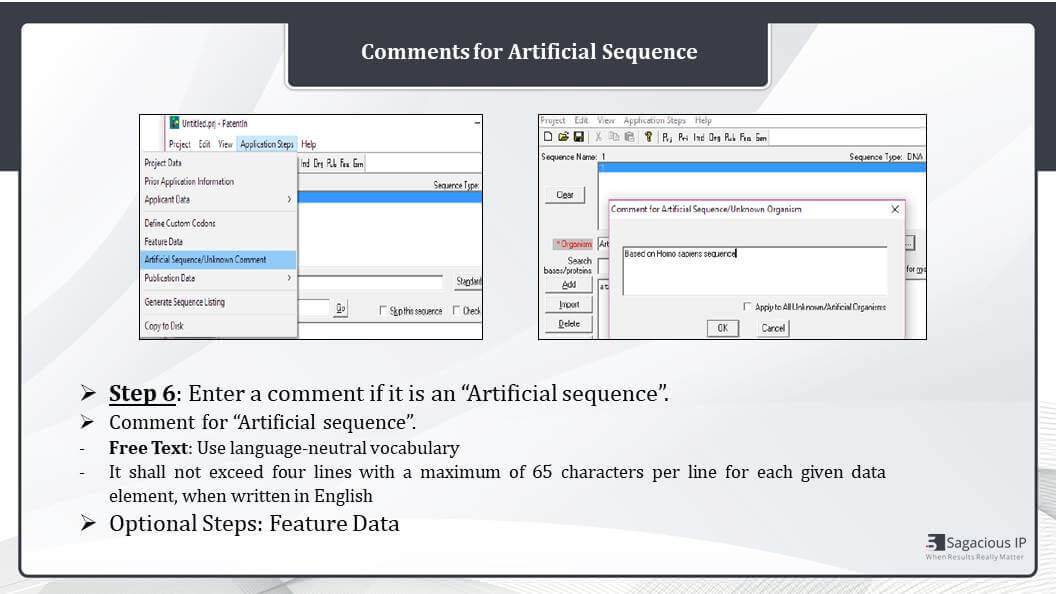
Another optional step is Feature Data that is entered like you can see the tab here, feature data. Details are entered in case of modified sequences.
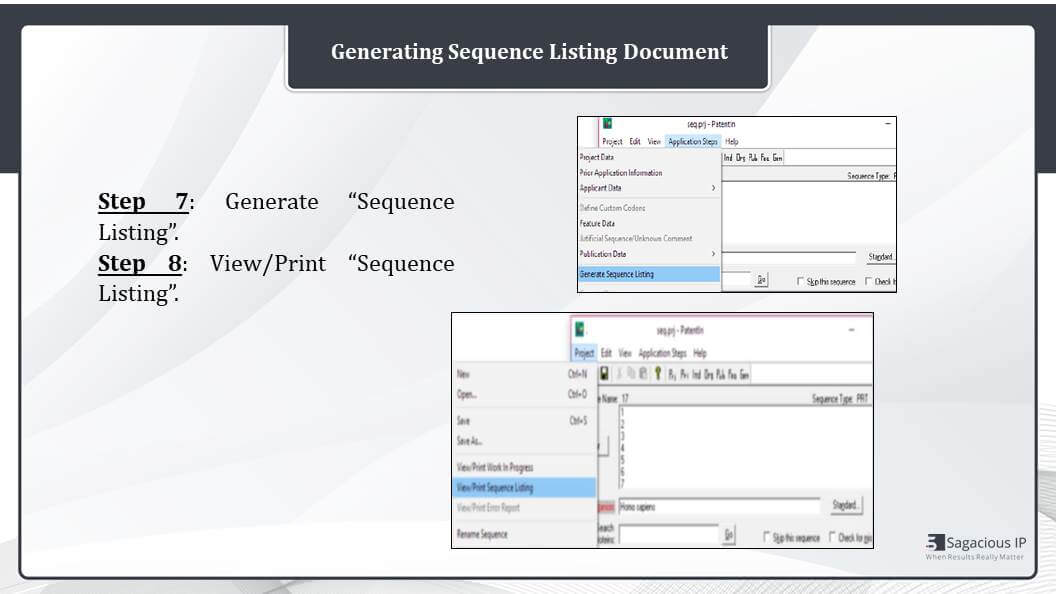
Generating Sequence Listing
The next step is the generating of sequence listing from the application steps bar. And once we generate the sequence listing, we can print it from the project tab here.
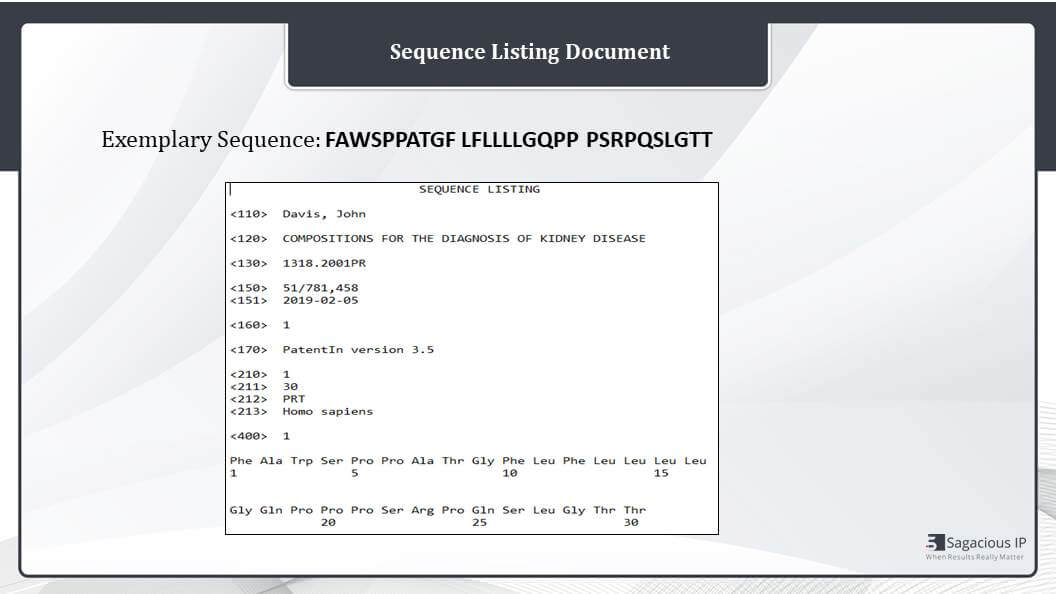
Sequence Listing Document
So this is how the final sequence listing document will look like. If this exemplary sequence is taken, then it will appear like this in the final sequence listing document. And if we see this Sequence Identifier 110, it stands for inventor, then 120 for the title of invention.
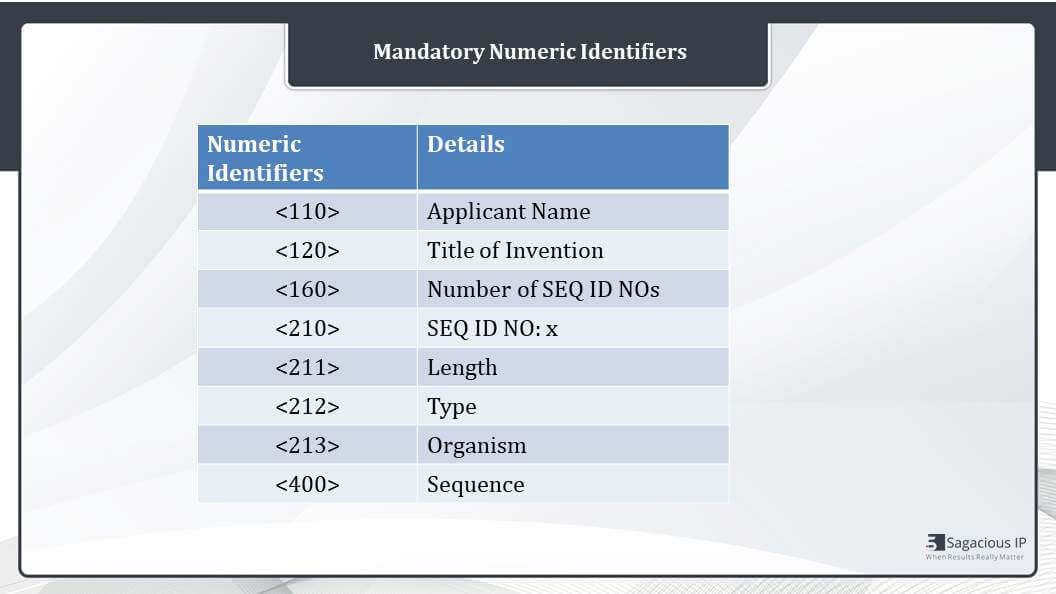
Mandatory Numeric Identifiers
If we see the numeric identifiers in details, here is the list, as I discussed, 110 for applicant name 120 for title of invention, then 160 that is number of SEQ ID NOs. If there is only one sequence, it is showing too. But if there were 30 sequences, the number would have been 30. Then 210 is the SEQ ID NO. 210 is 1 and similarly 211 length that is 30. It is a 30 amino acids sequence, so then 213 stands for organism and 400 for the sequence. You get the 400. It’s just showing the sequence of amino acids or protein sequences.
Conditional Numeric Identifiers
Apart from these, there are additional headers like 130. It is a file reference. It is provided when a sequence listing is filed at any time prior to the assignment of an application number. So a file reference is provided. Then, 140 and 141 are provided when we have the application details. And if the patent application is claiming priority to a prior application, then 150 and 151 should be filled.
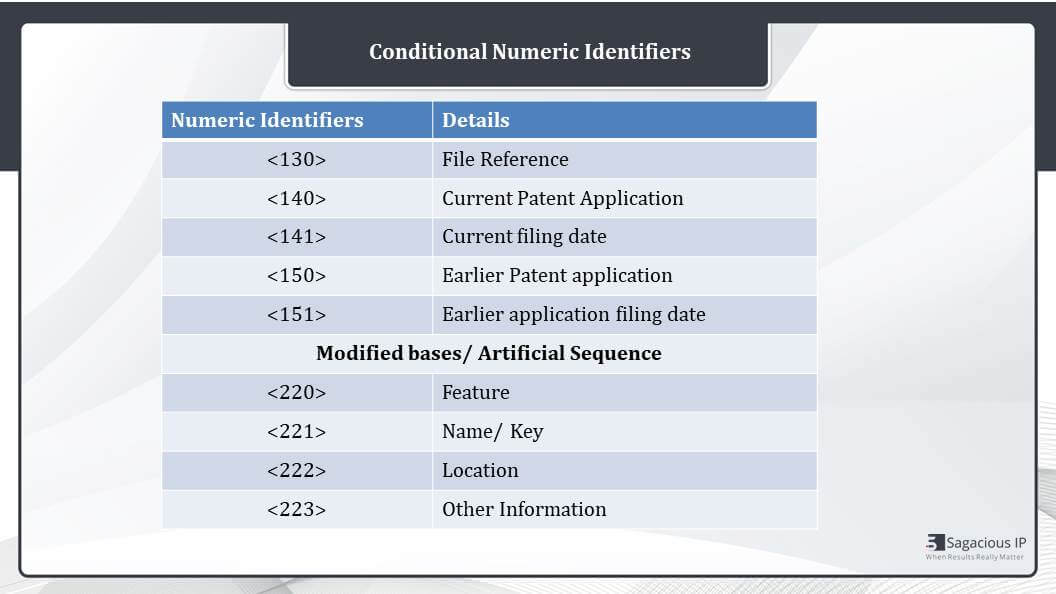
The next, in case of modified bases or artificial sequences, if an “N”or Xaa or modified base is used in the sequence, these four Sequence Identifiers are mandatory to be filled. There are Feature, Name/Key, Location, and Other information. And in case of unknown organism or artificial sequence 220 and 223 are mandatory.
Sequence Listings of Modified Sequences
Now let’s discuss the sequence listing for modified sequences. The biotechnology inventions don’t just talk about simple nucleotide sequences, but they consist of multiple modifications.
Types of Modified Sequences
If we read the sequence rules closely, it says all unbranched nucleotide sequences with 10 or more nucleotide bases and all unbranched, non-D amino acid sequences with four or more amino acids, provided that there are at least 10 “specifically defined” nucleotides or four specifically defined or amino acids, they should be listed. Here these “specifically defined” means the known nucleotides or amino acids.
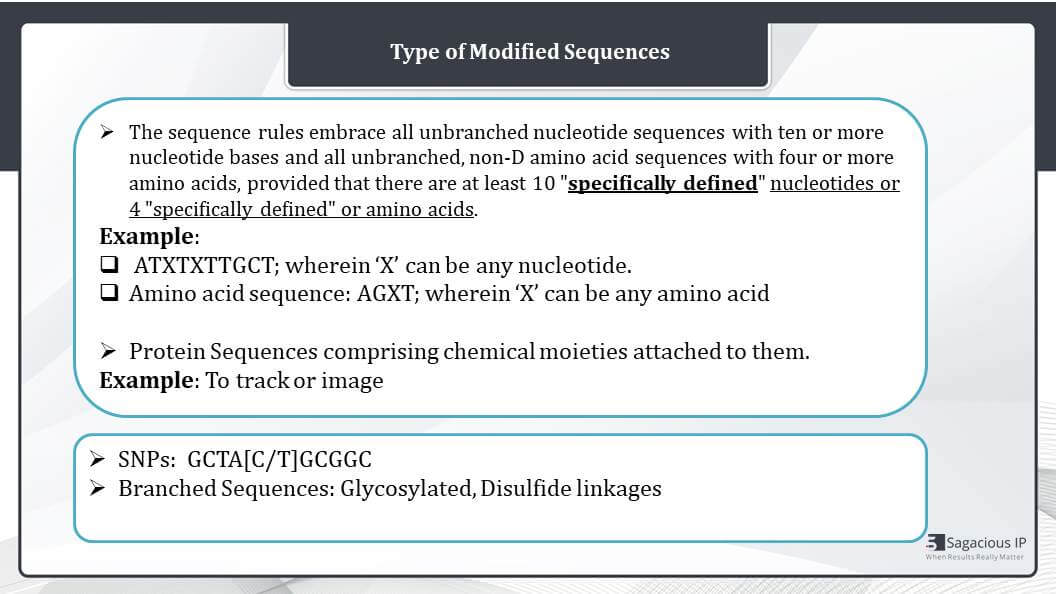
For example, we say this sequence ATXTXTTTGCT wherein X can be any nucleotide. Here it contains 10 nucleotide bases, so as per the rules, this should be listed. But only eight of them are specifically defined. Hence, such a sequence need not be listed because it does not have 10 specifically defined nucleotides. Similarly, in case of the next sequence, the amino acid sequence: AGXT, it contains only three specifically defined amino acids, so no need to list this in the sequence listing document.
Further, for protein sequences comprising chemical moieties for imaging purposes, only the protein sequence need to be listed and the chemical moiety information is to be provided in the patent application, not in a sequence listing document.
The other kind of modified sequences comprise single nucleotide polymorphisms, where there can be a difference of single nucleotide, then the other modified sequences can be glycosylated protein sequences, presence of Disulfide linkages or phosphorylated bases. And there are different rules to present different type of modifications. The more the modifications, the more different variety of rules need to be applied.
St 25 document provides a list of modified nucleotides and amino acids and below is an exemplary list. If the modified amino acid is 4-acetylcytidine, then it would be represented by the symbol given on the left, and same applies for the other examples. But if the modified nucleotide or amino acid does not fall in the previous list, then there are feature data keys to represent them.
Like here, we have to select the feature data, and then we have to provide a comment for the same in this position. So there are multiple variations for each kind of modifications.
If we see the representation for one of the feature data keys, here we can see the 221 Identifier shows CDS. And CDS is coding sequence that is sequence for nucleotides that corresponds with the sequence of amino acids in a protein. Here, it defines that it’s a coding sequence from 279 to 389 base pair. This is one kind of representation of modified sequences.
Case Studies
Now, further, let’s look at some of the case studies at Sagacious IP.
Case Study 1: How Sagacious Helped a US based Law Firm in Filing a PCT application comprising Biological Sequences
One of the cases, a US law form approached sagacious for sequence listing during the filing of a PCT application comprising biological sequences, and we used a PatentIn software version 3.5 for the preparation of the listing. Then, the nucleotide sequences also included modified bases. The modification details were mentioned in the comment section for artificial sequences and unknown organism as it was appropriate for the nucleotide.
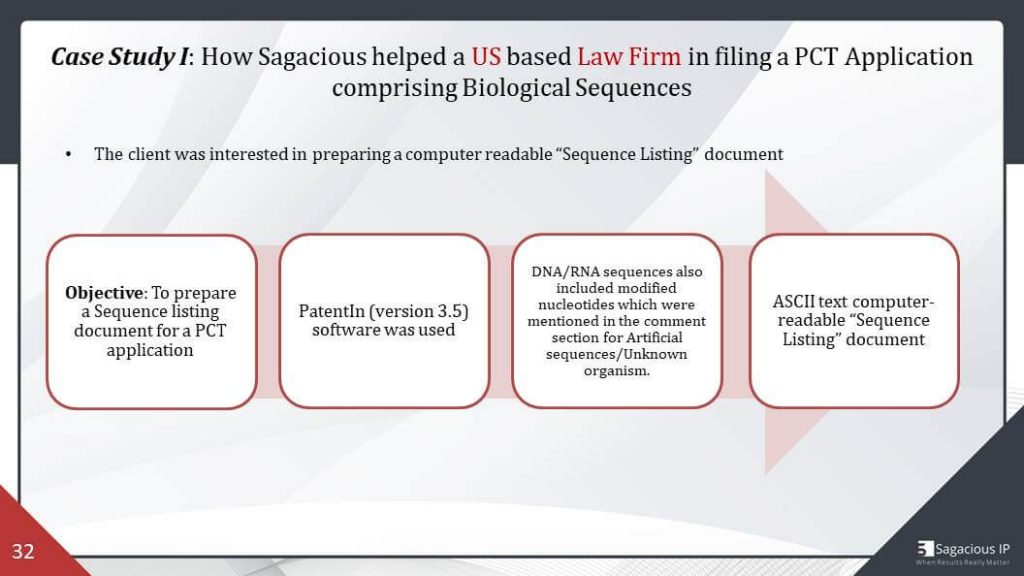
I remember it was some phosphorylated bases, so we mentioned the comments accordingly. Consequently, a computer readable sequence listing document was prepared, as per St25 standard, and it was submitted along with the patent application by the client.
Case Study 2: How Our Sequence Listing Service Helped a Europe Based Attorney to File a Corrected Sequence Listing after Office Action in USPTO.
In another case, a European based attorney approached Sagacious for the preparation of corrected version sequence listing.
An office action was received with respect to the sequence listings, and the client approached us. Unfortunately, he was not aware of the errors in the listings. So we first identified all the errors in the sequence listings and then we corrected them and prepared a modified and corrected version for these same sequences. Consequently, the corrected version was submitted to the USPTO.
So to conclude, we had discussed what sequence listing is, the importance and the advantages of sequence listings, then the key considerations to file a sequence listing, the timelines for filing the sequence listing document. Further, information needed for a sequence listing document. If you are going to do a sequence listing or you’re going to export to get sequence listing done, then the information required as the protein or nucleotide sequence organism name.
If it’s an artificial sequence, then there should be explanation for the same. Further, the patent application details, for example, applicant details, current and prior patent application details. And if there are modified sequences, then information needed would be the protein and amino acid sequences in the more modification details.
For example, the type of modification of the position of the nucleotide. I think this is mainly in the specifications, but if we are specifically going for sequence listing, we should keep this information separate.
Are there any questions?
Questions and Answers
Is There a Major Difference Between the Sequence Prepare under the CFR rules of US and St25 standard?
Manisha Rani – Pooja, I would like to ask a question if there is a major difference between the sequence prepare under the CFR rules of US and St25 standard.
Pooja Chhikara – Well, Manisha, no, there is no major difference in two of them. They are almost same and follow the same rules for presentation of sequences. It is just that USPTO defines CFR rules and St25 standards discuss more about the PCT application and its requirement details. Otherwise, the presentation and the sequence identifiers, they’re exactly same. They follow the same standard.
Manisha Rani – Okay, thank you for the explanation, Pooja!
Should I Get into Sequence Listing if I Have Two Sequences in Description?
We have another question from the audience. That is if someone is asking that I have two sequences, and that too in the description, should I get into sequence listing?
Here, I would like to answer. The answer is yes. There is no cap or the number of sequences and all type of sequences, whether claim or not, it should be listed. So according to already discussed standard format, that is CFR 1.821-1.825 or according to St25 format, all the nucleotide sequence more than 10 nucleotides and more than four amino acid needs to be listed.
Can I Do Sequence Listing by Myself?
Pooja Chhikara – Manisha, we have another question from the audience. It says I’m an inventor, and can I prepare the sequence listings myself, or should I approach the experts only?
Manisha Rani – Okay, here.
I would like to mention that anyone can do the sequence listing. But to get error free listing, it is advisable to reach out to the expert. As we discussed during the webinar, that biological invention contains simpler as well as modified biological sequences that are little too difficult to understand. However, if an inventor is doing the listing itself, it’s good to reach out to expert for final review.
Pooja Chhikara – Fair enough.
Thanks, Manisha.
I think the experts are the one who are dealing with sequence listings day in and day out, so it is always good to have their opinion about the same.
We have reached at the end of the webinar. This has been a wonderful session. I’m sure our listeners have great takeaways from this session and will be able to use several of these pointers when working on sequence listings. A few questions could not take up due to time limits. We hope to cover them through our subsequent write-up that we publish post this webinar. Thank you Manisha for joining us. The participants of this webinar can drop us an email at webinar at Sagacious Research com.
I want to extend a big thank you to our listeners who helped us start on time and finish as well. We highly appreciate that, and thank you very much.
Have a great day ahead.
Thank you again.
Submit Your Information to watch the Webinar Video:
"*" indicates required fields




